NATURE | NEWS
Replications, ridicule and a recluse: the controversy over NgAgo gene-editing intensifies
08 August 2016
SHIJIAZHUANG, CHINA
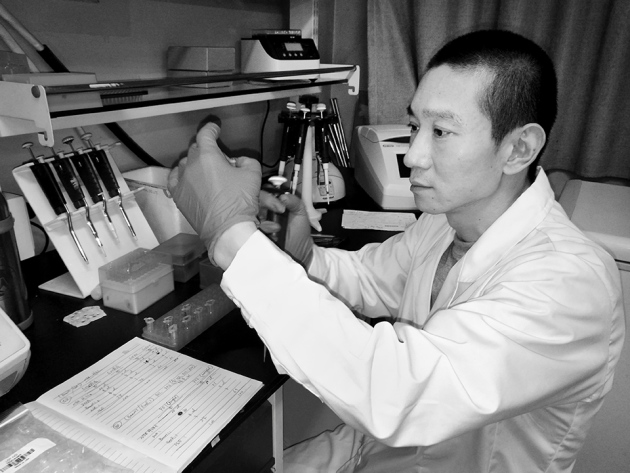
Biologist Han Chunyu receives dozens of harassing calls and texts each day, but he is convinced that his gene-editing paper is sound.
A controversy is escalating over whether a gene-editing technique proposed as an alternative to the popular CRISPR–Cas9 system actually works.
Three months ago, Han Chunyu, a biologist at Hebei University of Science and Technology in Shijiazhuang, reported that the enzyme NgAgo can be used to edit mammalian genes. Now, an increasing number of scientists are complaining that they cannot replicate Han’s results — although one has told Nature that he can.
Han says he receives dozens of harassing calls and texts each day, mocking him and telling him that his career is over — but he is convinced that the technique is sound. He also told Nature that he had submitted a detailed protocol to the online genetic-information repository Addgene on 8 August, at Addgene’s request, and hopes that this will help efforts to reproduce his work. Nature Biotechnology, which published the research, is investigating the matter.
The stakes are high. Over the past few years, the CRISPR–Cas9 system has transformed biology. But it has also made scientists hungry for other methods to expand the gene-editing toolkit: NgAgo is one of several that have emerged. “A lot of us are really cheerleading and hoping that it works,” says geneticist George Church of Harvard Medical School in Boston, Massachusetts.
CRISPR–Cas9 uses small genetic sequences to guide an enzyme to cut DNA in a particular location. Inspired, Han looked through the literature for other guidable protein ‘scissors’, and came across a family of proteins called Argonaute, or Ago, that fitted the bill. Others had flagged the proteins as potential gene editors.

Former biochemist Fang Shimin alleged on his website that the NgAgo paper was irreproducible.
In the paper, Han’s team reports using a wide variety of genetic sequences to guide one of these proteins, NgAgo, to edit eight different genes in human cells and to insert genes at specific points on chromosomes (F. Gao et al. Nature Biotechnol.34, 768–773; 2016).
Crucially, NgAgo very specifically cut only the target genes, says Han, unlike CRISPR–Cas9, which sometimes edits the wrong genes. And whereas CRISPR–Cas9 requires a certain genetic sequence to be near the cutting site to initiate its activity, NgAgo does not, which could broaden its potential applications, adds Han.
The initial reaction to the work in China was laudatory, including a visit to the lab by China Central Television. It was overwhelming, says Han, who is a reclusive figure. His hobbies include collecting teas and playing an ancient stringed instrument called the guqin. He doesn’t like to travel and has never left China: a trip to visit a collaborator in Hangzhou in March was the first time the 42 year old had boarded a plane. Before his paper came out, “I was completely unknown”, says Han, who spoke to Nature at his laboratory and a nearby restaurant.
Doubts about the research first surfaced at the beginning of July, when Fang Shimin, a former biochemist who has become famous for exposing fraudulent scientists, wrote on his website New Threads (xys.org) that he had heard reports of failed reproduction efforts, and alleged that Han’s paper was irreproducible. Criticism grew on various Chinese sites.

Geneticist Gaetan Burgio published failed attempts to replicate the NgAgo experiments on his blog.
On 29 July, the controversy went international when Gaetan Burgio, a geneticist at the Australian National University in Canberra, posted thorough details of his failed attempts to replicate the experiment on his blog. Normally, his posts get a few dozen hits, but this one spiked to more than 5,000.
On the same day, geneticist Lluís Montoliu, at the Spanish National Centre for Biotechnology in Madrid, e-mailed his colleagues at the International Society for Transgenic Technologies to recommend “abandoning any project involving the use of NgAgo” to “avoid wasting time, money, animals and people”. The e-mail was leaked and posted on Fang’s website.
Since then, an online survey by Pooran Dewari, a molecular biologist at the MRC Centre for Regenerative Medicine in Edinburgh, UK, has found only 9 researchers who say that NgAgo works— and 97 who say that it doesn’t.
Two researchers who initially reported success with NgAgo in an online chat group now say that they were mistaken. Debojyoti Chakraborty, a molecular biologist at the CSIR-Institute of Genomics and Integrative Biology in New Delhi, says that he repeated a specific section of Han’s paper that described using NgAgo to knock out a gene for a fluorescent protein that had been introduced into a cell. The glow was reduced, so Chakraborty assumed that NgAgo had disabled the gene. But after sequencing the DNA, he found no evidence of gene editing. He now says that the reduction in fluorescence must have had some other cause.
Jan Winter, a PhD student in genomics at the German Cancer Research Center in Heidelberg, says that he had a similar experience. “I will retry the experiment in the upcoming weeks, but so far I think it won’t work,” he says.

Geneticist Lluís Montoliu recommends abandoning any project involving the use of NgAgo for gene-editing.
Han says that he has only got the system to work on cells cultured in his laboratory, and it failed in cells that he purchased. He later found the purchased cells to be contaminated with bacteria called mycoplasma, and says that others might be having the same problem. He adds that some graduate students might be working too fast and not being careful with reagents. Winter disagrees: “I do not think it is a problem of the scientists doing something wrong.”
One researcher in China, who works independently from Han’s research group and who doesn’t want his name to be entangled in the public controversy, told Nature that he had tested NgAgo in a few kinds of cell and found that it was able to induce genetic mutations at the desired sites — a finding that he verified by sequencing. He adds that the process was less efficient than CRISPR–Cas9, and requires tweaking to improve the efficiency. “But, in short, it worked,” he says.
Two more Chinese scientists, who also asked not to be named, say they have initial results showing that NgAgo works but still need to confirm with sequencing.
“It might, might work,” says Burgio, “but if so, it’s so challenging that it’s not worth pursuing. It won’t surpass CRISPR, not by a long shot.”
The failure of NgAgo “would be disappointing, but then there is work for us left to do to see whether other Argonaute systems can get it to work somehow,” says microbiologist John Van der Oost of Wageningen University in the Netherlands, a co-author of the 2014 analysis of Argonaute proteins that laid the groundwork for their use in gene editing (D. C. Swaarts et al. Nature 507, 258–261; 2014).
This week, Nature Biotechnology sent a statement to Nature’s news team, saying that “several researchers” have contacted the journal to report that they cannot reproduce the results, and that “the journal is following established process to investigate the issues”. A spokesperson declined to comment on the nature or duration of the investigation. (Nature Biotechnology is published byNature’s publisher, Springer Nature; Nature’s news and comment team is editorially independent of the publisher’s research editorial teams.)
Hebei University says that it will ask Han to repeat the experiment so that it can be verified by an independent party within a month, according to Chinese state media.
Nature
536,
136–137
(11 August 2016)
Replications, ridicule and a recluse: the controversy over NgAgo gene-editing intensifies
08 August 2016
SHIJIAZHUANG, CHINA

Biologist Han Chunyu receives dozens of harassing calls and texts each day, but he is convinced that his gene-editing paper is sound.
A controversy is escalating over whether a gene-editing technique proposed as an alternative to the popular CRISPR–Cas9 system actually works.
Three months ago, Han Chunyu, a biologist at Hebei University of Science and Technology in Shijiazhuang, reported that the enzyme NgAgo can be used to edit mammalian genes. Now, an increasing number of scientists are complaining that they cannot replicate Han’s results — although one has told Nature that he can.
Han says he receives dozens of harassing calls and texts each day, mocking him and telling him that his career is over — but he is convinced that the technique is sound. He also told Nature that he had submitted a detailed protocol to the online genetic-information repository Addgene on 8 August, at Addgene’s request, and hopes that this will help efforts to reproduce his work. Nature Biotechnology, which published the research, is investigating the matter.
The stakes are high. Over the past few years, the CRISPR–Cas9 system has transformed biology. But it has also made scientists hungry for other methods to expand the gene-editing toolkit: NgAgo is one of several that have emerged. “A lot of us are really cheerleading and hoping that it works,” says geneticist George Church of Harvard Medical School in Boston, Massachusetts.
CRISPR–Cas9 uses small genetic sequences to guide an enzyme to cut DNA in a particular location. Inspired, Han looked through the literature for other guidable protein ‘scissors’, and came across a family of proteins called Argonaute, or Ago, that fitted the bill. Others had flagged the proteins as potential gene editors.

Former biochemist Fang Shimin alleged on his website that the NgAgo paper was irreproducible.
In the paper, Han’s team reports using a wide variety of genetic sequences to guide one of these proteins, NgAgo, to edit eight different genes in human cells and to insert genes at specific points on chromosomes (F. Gao et al. Nature Biotechnol.34, 768–773; 2016).
Crucially, NgAgo very specifically cut only the target genes, says Han, unlike CRISPR–Cas9, which sometimes edits the wrong genes. And whereas CRISPR–Cas9 requires a certain genetic sequence to be near the cutting site to initiate its activity, NgAgo does not, which could broaden its potential applications, adds Han.
The initial reaction to the work in China was laudatory, including a visit to the lab by China Central Television. It was overwhelming, says Han, who is a reclusive figure. His hobbies include collecting teas and playing an ancient stringed instrument called the guqin. He doesn’t like to travel and has never left China: a trip to visit a collaborator in Hangzhou in March was the first time the 42 year old had boarded a plane. Before his paper came out, “I was completely unknown”, says Han, who spoke to Nature at his laboratory and a nearby restaurant.
Doubts about the research first surfaced at the beginning of July, when Fang Shimin, a former biochemist who has become famous for exposing fraudulent scientists, wrote on his website New Threads (xys.org) that he had heard reports of failed reproduction efforts, and alleged that Han’s paper was irreproducible. Criticism grew on various Chinese sites.

Geneticist Gaetan Burgio published failed attempts to replicate the NgAgo experiments on his blog.
On 29 July, the controversy went international when Gaetan Burgio, a geneticist at the Australian National University in Canberra, posted thorough details of his failed attempts to replicate the experiment on his blog. Normally, his posts get a few dozen hits, but this one spiked to more than 5,000.
On the same day, geneticist Lluís Montoliu, at the Spanish National Centre for Biotechnology in Madrid, e-mailed his colleagues at the International Society for Transgenic Technologies to recommend “abandoning any project involving the use of NgAgo” to “avoid wasting time, money, animals and people”. The e-mail was leaked and posted on Fang’s website.
Since then, an online survey by Pooran Dewari, a molecular biologist at the MRC Centre for Regenerative Medicine in Edinburgh, UK, has found only 9 researchers who say that NgAgo works— and 97 who say that it doesn’t.
Two researchers who initially reported success with NgAgo in an online chat group now say that they were mistaken. Debojyoti Chakraborty, a molecular biologist at the CSIR-Institute of Genomics and Integrative Biology in New Delhi, says that he repeated a specific section of Han’s paper that described using NgAgo to knock out a gene for a fluorescent protein that had been introduced into a cell. The glow was reduced, so Chakraborty assumed that NgAgo had disabled the gene. But after sequencing the DNA, he found no evidence of gene editing. He now says that the reduction in fluorescence must have had some other cause.
Jan Winter, a PhD student in genomics at the German Cancer Research Center in Heidelberg, says that he had a similar experience. “I will retry the experiment in the upcoming weeks, but so far I think it won’t work,” he says.

Geneticist Lluís Montoliu recommends abandoning any project involving the use of NgAgo for gene-editing.
Han says that he has only got the system to work on cells cultured in his laboratory, and it failed in cells that he purchased. He later found the purchased cells to be contaminated with bacteria called mycoplasma, and says that others might be having the same problem. He adds that some graduate students might be working too fast and not being careful with reagents. Winter disagrees: “I do not think it is a problem of the scientists doing something wrong.”
One researcher in China, who works independently from Han’s research group and who doesn’t want his name to be entangled in the public controversy, told Nature that he had tested NgAgo in a few kinds of cell and found that it was able to induce genetic mutations at the desired sites — a finding that he verified by sequencing. He adds that the process was less efficient than CRISPR–Cas9, and requires tweaking to improve the efficiency. “But, in short, it worked,” he says.
Two more Chinese scientists, who also asked not to be named, say they have initial results showing that NgAgo works but still need to confirm with sequencing.
“It might, might work,” says Burgio, “but if so, it’s so challenging that it’s not worth pursuing. It won’t surpass CRISPR, not by a long shot.”
The failure of NgAgo “would be disappointing, but then there is work for us left to do to see whether other Argonaute systems can get it to work somehow,” says microbiologist John Van der Oost of Wageningen University in the Netherlands, a co-author of the 2014 analysis of Argonaute proteins that laid the groundwork for their use in gene editing (D. C. Swaarts et al. Nature 507, 258–261; 2014).
This week, Nature Biotechnology sent a statement to Nature’s news team, saying that “several researchers” have contacted the journal to report that they cannot reproduce the results, and that “the journal is following established process to investigate the issues”. A spokesperson declined to comment on the nature or duration of the investigation. (Nature Biotechnology is published byNature’s publisher, Springer Nature; Nature’s news and comment team is editorially independent of the publisher’s research editorial teams.)
Hebei University says that it will ask Han to repeat the experiment so that it can be verified by an independent party within a month, according to Chinese state media.
Nature
536,
136–137
(11 August 2016)





